|
|
“THIS IS AN ANIMAL that lives inside the anus of sea cucumbers,” says Giacomo Bernardi, holding an oblong bottle at eye level to examine the white, 10-inch-long, snakelike specimen inside. Bernardi—a tanned, balding scientist clad in a sweatshirt, shorts, and sneakers—is showing off the fish he has collected around the world, from the anal albino inhabitant to colorful coral reef dwellers.
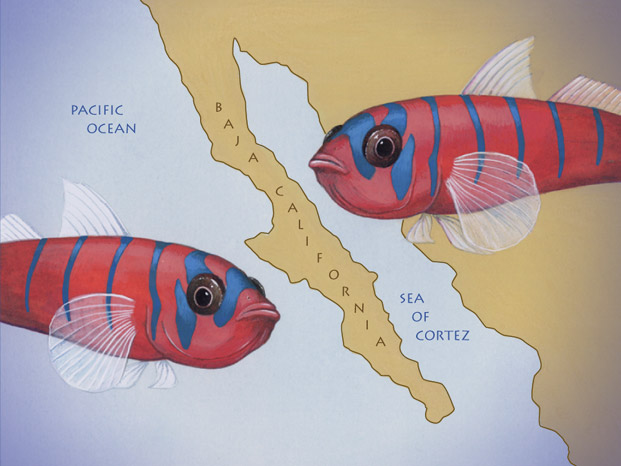
Jars of pickled fish line the shelves in Bernardi’s office and the countertops in his lab at the University of California's Long Marine Laboratory in Santa Cruz. Bernardi moves to a refrigerator, opens the door, and produces a clear plastic tube. Inside, several quarter-sized tropical damselfish float in alcohol. These adult fish are black with a small white dot near the eye, the remnants of a white streak that fades with maturity, Bernardi explains. He then opens one of many boxes to reveal rows of vials—each one encasing a miniature, M&M-sized juvenile damselfish, white stripe and all. Although these critters look identical, a peek at DNA harvested from their gills can reveal a surprise: that a group appearing to comprise only one type of fish might actually harbor several species. Bernardi is an evolutionary biologist who ponders marine creatures for the same reason Darwin scrutinized finches: to learn what drives the diversity of life. “There are 25,000 species of fish, which is more than all the land vertebrates put together,” Bernardi says. “But people don’t exactly understand how species of fish are created.” Gaining insights into that process could help scientists estimate the variety of fish in the sea. These estimates are important because the number of species and the abundance of each animal help scientists decide which ocean zones deserve protection. By analyzing tiny differences in DNA, Bernardi can determine how closely related members of a given species are—and even detect when some members have begun to split off into new subspecies. “Our tools allow us to uncover things you can’t really see,” he says. Indeed, Bernardi has recently captured molecular snapshots of evolution in action among fish swimming in the waters off the Baja California peninsula. His laboratory trove includes a haul of sandbass, rockfish, and other marine creatures. DNA analyses of the specimens suggest that some types of fish began forming new species when the Baja peninsula rose from the ocean millions of years ago, dividing a single population into two distinct groups, one on each side of the wall formed by the peninsula. The research is a prime new example of how geologic events can produce new species—and also underscores the fact that the ocean harbors far more variety in its denizens than anyone realized. CATALOGUING THE PLANET’S inhabitants is the mother of all census projects. Scientists have so far documented nearly 2 million species of bacteria, plants, fungi, and animals. Yet the globe hosts 5 to 30 million species of organisms, biologists estimate. Every time scientists discover new life forms, taxonomists toil to properly categorize the creatures. Part of the challenge lies in the definition of a species—there is no hard-and-fast rule. Looks and lifestyle can offer a clue; animals that generally look alike, live in a similar habitat, and prey on the same food source are typically classified together. But comparable appearances and lifestyles can be deceiving. For example, sharks and dolphins are both large, finned swimmers that eat smaller fish, yet sharks are a type of fish and dolphins are a type of mammal. The two groups have completely separate ancestral lines. Among animals that are closely related, the task of categorizing species can be even more daunting. Creatures that look and act similarly sometimes belong to different classification groups—consider the 450 species of North American ladybugs. Conversely, one group can comprise many creatures that seem very different from one another. Rottweilers and poodles, for instance, are members of the same species. Mating success or failure can additionally help scientists to properly classify animals; a species is often defined by the ability of members to mate and produce fertile offspring. Yet biologists cannot always assess breeding feasibility, especially when different populations of the same species don’t normally encounter each other in nature. For example, the bottlenose dolphin species contains Atlantic and Pacific varieties, yet it’s unknown whether the two groups can actually breed with each other. While such questions may have confounded Darwin’s peers, modern scientists have an advantage—they can scrutinize the molecular makeup of organisms for fresh insights. The more genetic differences that exist between animals within a species, the more likely it is that the population has split into subspecies that can no longer interbreed. Such subgroups are most likely to form within a "disjunct" species—a species whose populations have been geographically separated from each other into two discrete regions. The human species experienced small-scale disjunction, for instance, when the demilitarized zone of Germany's Berlin Wall divided families for three decades. On the evolutionary scale, barriers such as mountains, oceans, or simply large distances can suddenly split land-dwelling populations. Such obstacles are so vast and permanent that disjunct creatures sometimes eventually evolve into different subspecies. In the sea, land masses, water temperatures, or ecological niches such as reefs or tide pools can corral marine life. One well-studied barrier is the Isthmus of Panama, connecting North and South America. When this strip of land jutted out of the ocean 3.5 million years ago, it divided populations into Pacific and Caribbean groups. Today, several species are found on both sides of the divide. Yet biologists have detected molecular changes revealing that a number of those disjunct species have began to diverge into distinct subgroups. HOPING TO DISCOVER more emerging subspecies, Bernardi recently headed to Baja California, a region chock-full of known disjunct species that have not yet been studied on a molecular level. The Baja peninsula is a narrow land strip extending 800 miles south of California. The Pacific Ocean laps at its western shores, while on the east, the Sea of Cortez separates the peninsula from mainland Mexico. The waters surrounding the peninsula teem with fish, but only a handful of species are sharply divided by the geography. Those species reside in the cool water around the northern portion of Baja California, and two barriers fence them in: the land itself and warm southern waters. Baja California represents one of the rare regions where genetic surveys of disjunct species are possible. “There are very few places where you have all the good things to study—[where] you can sample the species, you know what they are, you know what the currents are, you know the geologic history of the place, you happen to find an abundance of individuals,” Bernardi says. So Bernardi braved chilly waters at 19 locations up and down both sides of Baja California to gather his gilled guinea pigs. Sometimes he used a snorkel and a hand-held net, other times he speared his quarry while scuba diving. Occasionally he needed only to wade in tide pools. Bernardi collected fish representing nine disjunct species, including variations of sandbass, rockfish, perch, and grunion. He nabbed anywhere from one to 44 fish at each site and hauled more than 200 pickled specimens back to his laboratory for genetic analysis. After harvesting DNA from the fish he’d collected, Bernardi compared the genetic sequences of individual fish within a single species. Although genetic makeup is bound to vary among individuals of one species, more than 99 percent of their DNA is estimated to be identical. The amount of DNA variability among organisms is a little like the differences and similarities among automobiles. Non-essentials such as color and carpeting vary widely among cars, just as genes for coloring or personality differ in individual animals. More fundamental aspects such as engine and frame design will be common to a particular class of car; likewise, related species share many fundamental genes, such as those dictating size and shape. And essentials like pistons and tires vary little, just as certain genes are virtually identical from mice to men. To effectively study the differences between his disjunct fish, Bernardi needed to focus on a genetic region known to foster just the right amount of variability. He chose a narrow segment of DNA called the D-loop control region. The D-loop resides within the mitochondria, the energy generators of the cell that harbor their own miniature set of genes. But the D-loop itself doesn’t encode a gene—it instead serves as sort of genetic filler. So, in the same way that the upholstery inside a car doesn’t affect its performance, random mutations within this stretch don’t help or hinder the survival of a fish species through the forces of natural selection. As a result, D-loop DNA changes are less likely to sicken or kill an animal (and thus more likely to be passed to the next generation) than mutations in sections that encode genes. An average animal gene might accumulate eight mutations over a million years, whereas the D-loop could chalk up as many as 40. Sequences of D-loop DNA are similar among animals that are breeding and exchanging genes with each other, but genetically isolated populations rack up differences quickly on an evolutionary timescale. So this rapidly changing region serves as a sort of molecular clock: The greater the number of DNA differences, the longer it has been since the groups have diverged. FOR EACH of the nine disjunct Baja species, Bernardi scrutinized and compared the DNA of at least five fish from the Pacific side of the Baja peninsula and five that had swum in the Sea of Cortez. A computer analysis of their genetic differences helped him rank how closely related the fish in each species were to each other. This information then allowed Bernardi to build a kind of evolutionary family tree, which clusters the fish into branches according to their similarities. His results show a genetic divergence brewing within several of the groups. For example, the evolutionary tree he generated for the grunion species showed a rift was forming between fish from the Pacific Ocean and those from the Sea of Cortez: The grunion living on one side of the peninsula were more closely related to each other than to grunion from the other side. He saw a similar pattern in two-thirds of the species overall. “The population used to be continuous, and then the geologic formation of Baja California physically separated them. And then they slowly diverged genetically,” Bernardi explains. For some of these diverging species, the number of DNA discrepancies suggests that the animals have been accumulating differences for about 2.5 million years—dating back to around the same period when the Baja peninsula formed. The data also hint that other species started branching apart less than 1 million years ago, roughly the time when an inland seaway closed, preventing marine life from crossing the peninsula. Bernardi’s findings suggest that the presence of the Baja peninsula is in fact causing new species to form. At the same time, Bernardi found that four of the species he studied are genetically uniform across the Baja; those species don’t seem to be splitting. That finding suggests that fish in those groups aren't truly separated and might be migrating around the southern tip of the peninsula. Alternatively, the uniform distribution could simply mean that those populations haven’t been separated long enough to accumulate DNA differences. Bernardi wonders if recent glacial events, which temporarily chilled the ocean about 10,000 years ago, could account for the consistency of these animals across the peninsula. A cooling of the seas surrounding the southern peninsula would allow migration of the cold-water fish, which would be restricted again when the waters warmed back up. Because separations occurring less than 150,000 years ago are virtually invisible to the genetic technique, Bernardi can’t distinguish recent splits from ongoing migrations. Whatever the explanation, Bernardi’s work paints a clearer picture of the diversity of fish around Baja California. Although he is not petitioning to reclassify the Baja’s eastern grunion in a different species from its western grunion, he says it is important to realize that the two populations are not homogeneous. “We have to acknowledge the fact that we’re talking about two general groups that are separated from each other—not only in space but also in time, by 1 million years,” he says. YET IT IS THE DISTANCE in time that worries some evolutionary biologists, who consider molecular clock analyses troublesome. Ideally, the DNA region chosen as a molecular clock (the D-loop, in Bernardi’s case) incurs random mutations that don’t affect the animal’s survival. Any mutation that confers a detriment or benefit would skew the timepiece. But researchers disagree over which regions of DNA, if any, accumulate harmless mutations regularly over time. “One of the problems is that people who use these molecular data as clocks have to make a number of assumptions,” says William Fink, an evolutionary biologist at the University of Michigan in Ann Arbor. “What they’re assuming is that the molecules they’re looking at are basically evolving randomly.... That’s hard to be sure about.” Also, DNA changes are not as predictable as the ticks of a clock—mutations do not necessarily occur once every so-many years. “It doesn’t always change regularly with time,” says geneticist Carol Stepien of Cleveland State University in Ohio. “That gives you slop in your clock.” But, she adds, the technique is widely accepted among evolutionary biologists, and she herself uses it. “You can tell a very interesting story if you use a molecular clock.” Fink and Stepien both agree, however, that scientists can bolster this kind of data by verifying how fast their clock ticks. One way to do that is to compare the estimated dates of species-branching events with those of known geologic events, such as creation of the Isthmus of Panama. If the molecular clock predicts that the branching happened around the same time as the shift in geology, researchers can be more confident that their timepiece ticks accurately. “When groups are separated on either side of the isthmus, we know when the door closed,” says Stepien. “When we have a clear separation of groups, we can calibrate a molecular clock.” In the case of Bernardi’s disjunct Baja fish, one lucky finding enables an extra measure of calibration. Unlike the Isthmus of Panama, the Baja peninsula is more like a door left ajar—it formed a “leaky” barrier that did not completely separate marine populations. This leakiness makes it more difficult for Bernardi to establish a firm timeline when using DNA mutations to estimate the years gone by. Bernardi would need to compare D-loop mutations among fish separated by a firmly closed door to calibrate the tickings of his molecular clock. It just so happens that one type of fish that’s forming subspecies across the Baja peninsula, the sargo, has cousins that were separated by the Isthmus of Panama. This rare find—two closely related disjunct species—enables Bernardi to compare his estimates of mutation rates to two geologic events. With his precisely calibrated molecular clock, Bernardi is confident that formation of the Baja peninsula triggered subspecies evolution. SUCH INVESTIGATIONS ALLOW scientists to peer beneath the sea’s surface and take stock of what’s living there. “We have a system that allows us to understand how new species are being created,” Bernardi says. “If you don’t understand that, it’s really difficult to understand how biodiversity is generated in the ocean.” Understanding aquatic diversity is critical when scientists decide the boundaries of federally protected marine reserves. Conservation goals can change when scientists learn that one species of, say, rockfish, is in fact two populations that no longer exchange genes with each other. “We use that [type of] info to try to decide how we want to manage those areas,” says Steve Palumbi, a biologist at Stanford University’s Hopkins Marine Station in Pacific Grove, California. “If there’s plenty of gene flow, we would manage in a different way than if that gene flow were impossible.” In a region such as the northern Baja Peninsula, for instance, if biologists worked toward conserving critters on only one coast, they would be neglecting the closely related, yet distinct subspecies on the other side. Research such as Bernardi’s gives evolutionary biologists a glimpse into how and when new species begin to form. “It provides a great road map into the past,” says Palumbi. The study of animal species on a molecular level offers modern scientists a window into evolution that Darwin would envy. Bernardi, for one, does not take this power lightly. In fact, it sometimes leaves him feeling like he’s discovering evolution all over again. Says Bernardi with a wink, “I am Darwin ... reincarnated!” |